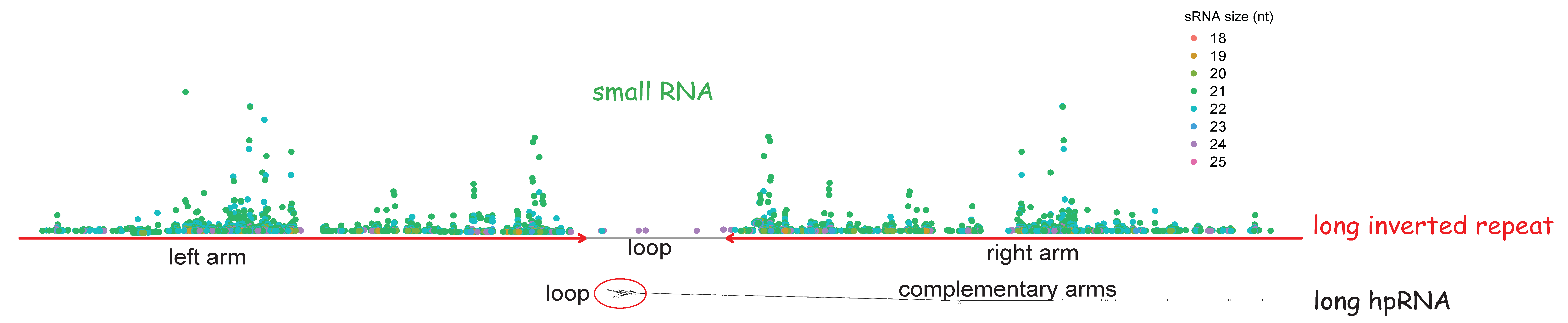
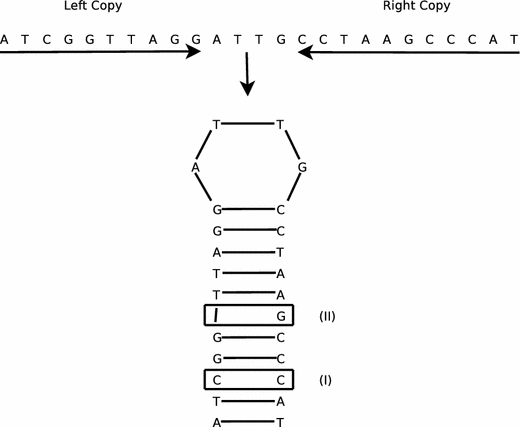
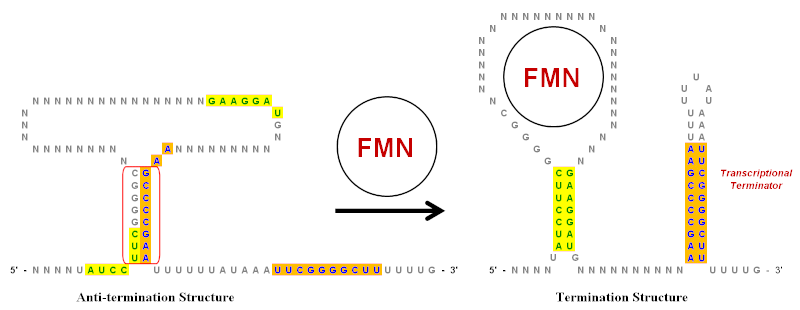
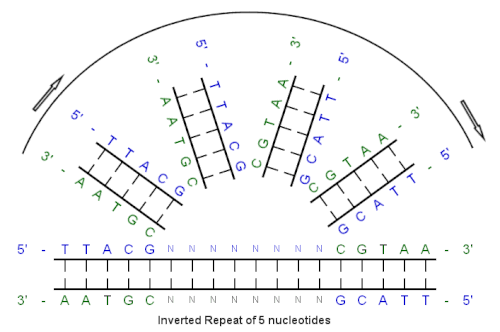
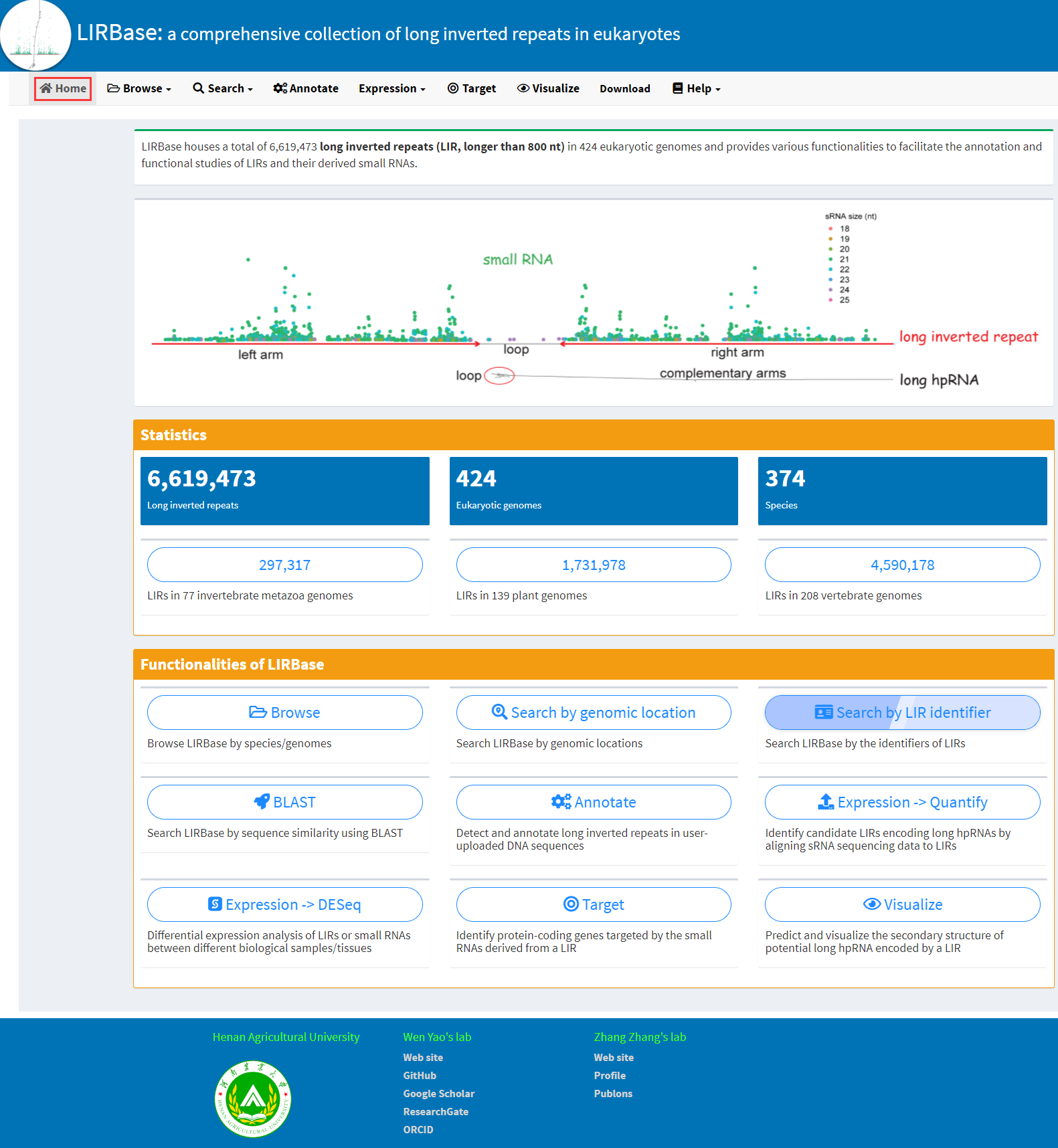
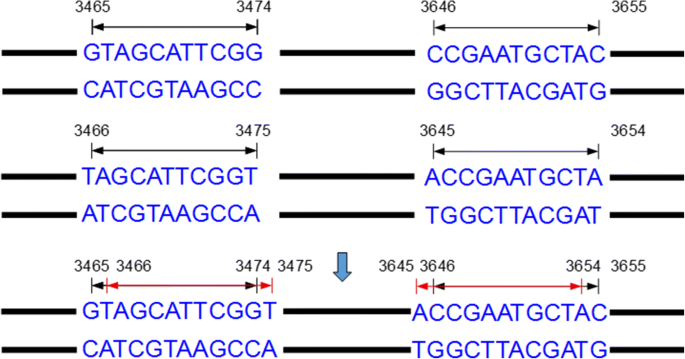
MUST: A system for identification of miniature inverted-repeat transposable elements and applications to Anabaena variabilis and Haloquadratum walsbyi - ScienceDirect
MiteFinderII: a novel tool to identify miniature inverted-repeat transposable elements hidden in eukaryotic genomes | BMC Medical Genomics | Full Text
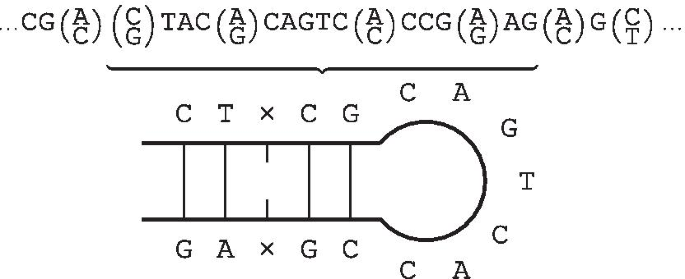
IUPACpal: efficient identification of inverted repeats in IUPAC-encoded DNA sequences | BMC Bioinformatics | Full Text
![PDF] detectIR: A Novel Program for Detecting Perfect and Imperfect Inverted Repeats Using Complex Numbers and Vector Calculation | Semantic Scholar PDF] detectIR: A Novel Program for Detecting Perfect and Imperfect Inverted Repeats Using Complex Numbers and Vector Calculation | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/a20f6bea6d62205d58f8bcd56fa3411c859aad47/3-Figure1-1.png)
PDF] detectIR: A Novel Program for Detecting Perfect and Imperfect Inverted Repeats Using Complex Numbers and Vector Calculation | Semantic Scholar

Comparative plastomics of Amaryllidaceae: Inverted repeat expansion and the degradation of the ndh genes in Strumaria truncata Jacq | bioRxiv
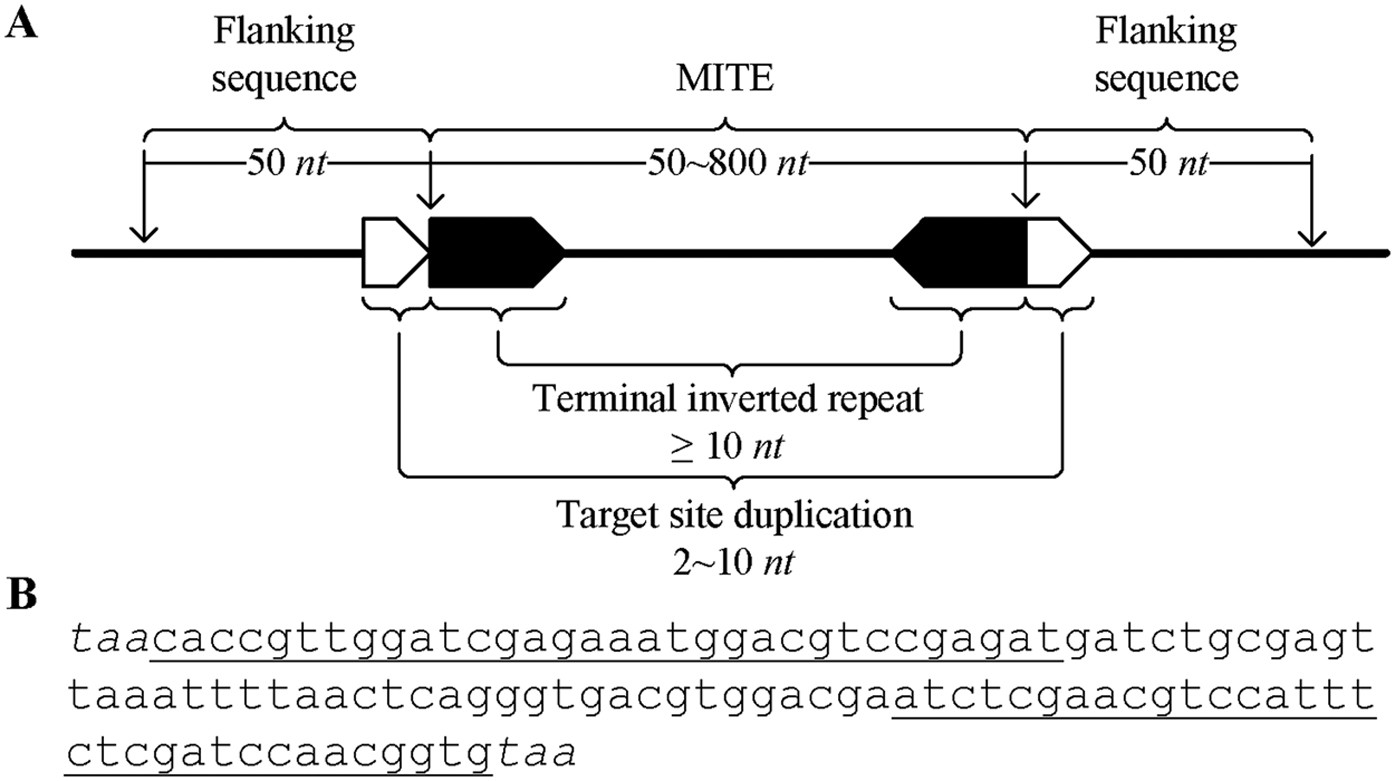
detectMITE: A novel approach to detect miniature inverted repeat transposable elements in genomes | Scientific Reports
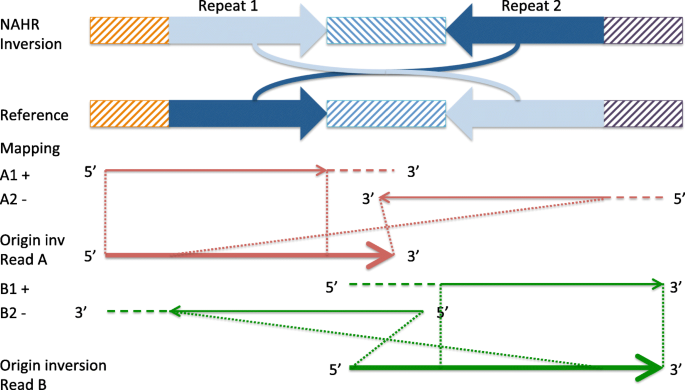
npInv: accurate detection and genotyping of inversions using long read sub-alignment | BMC Bioinformatics | Full Text

The short inverted repeats-induced circEXOC6B inhibits prostate cancer metastasis by enhancing the binding of RBMS1 and HuR: Molecular Therapy
detectIR: A Novel Program for Detecting Perfect and Imperfect Inverted Repeats Using Complex Numbers and Vector Calculation | PLOS ONE

Large inverted repeats identified by intra-specific comparison of mitochondrial genomes provide insights into the evolution of Agrocybe aegerita - ScienceDirect

Inverted repeat motif as a potential origin of replication. Sequences... | Download Scientific Diagram

Correlations between long inverted repeat (LIR) features, deletion size and distance from breakpoint in human gross gene deletions | Scientific Reports
![Comparative plastomics of Amaryllidaceae: inverted repeat expansion and the degradation of the ndh genes in Strumaria truncata Jacq. [PeerJ] Comparative plastomics of Amaryllidaceae: inverted repeat expansion and the degradation of the ndh genes in Strumaria truncata Jacq. [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2021/12400/1/fig-1-full.png)